Digging Deeper Into a Promethease Finding Before Accepting It as Truth
I recently worked with a client who was adopted and had used a raw data file and the Promethease tool as an avenue to obtain some health/medical information for herself.
This case gets a little complicated, but please stick with it; it demonstrates the areas where we discover weakness in our understanding and communication of medical genetics data.
The case also highlights the importance of doing a deeper investigation of findings that show up on a Promethease report, before accepting the report’s summary at face value. I write about 23andMe and Promethease in the summary, but my conclusion is true for any tool (Genetic Genie, Sequencing.com reports, etc.) run on any raw data file (AncestryDNA, MyHeritage, etc).
I could write up more cases where other companies’ raw data files analyzed by other third-party sites showed weaknesses as well, and I will try to do that in the future. For now, I am focusing on this one case to highlight major issues we are still facing and needing to address in the world of medical genetics.
The client came to me with her Promethease report already generated, having used a raw data file from 23andMe. Her concern was for a possibly elevated risk for gastric cancer based on a finding that appeared at the top of her report.
I applied my custom filtering process to the Promethease report to narrow down the findings to a manageable size (here is the post describing how and why I do that).
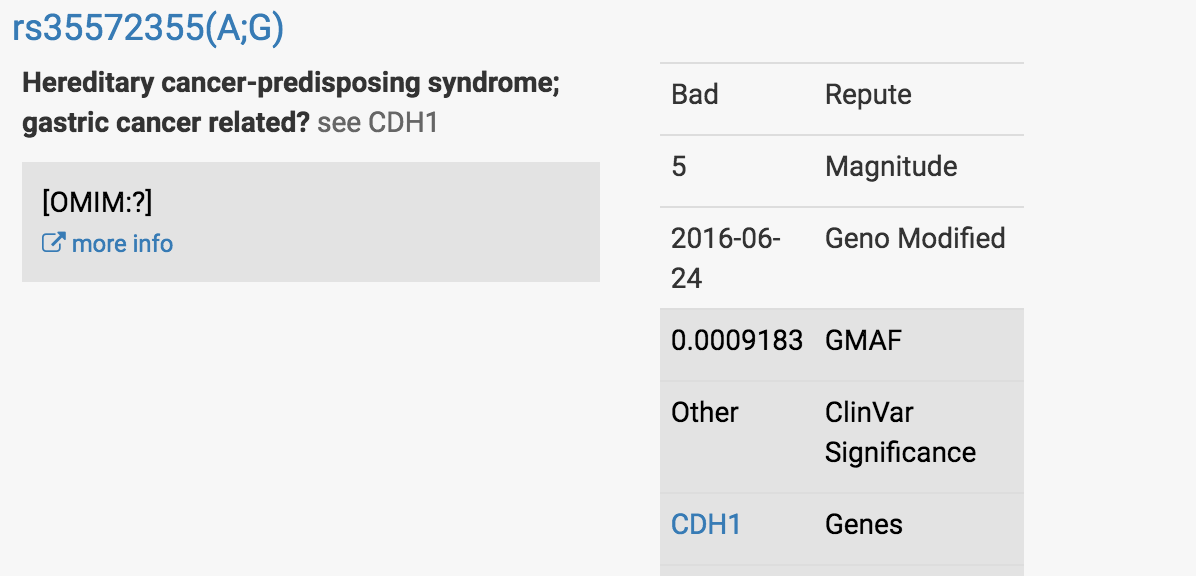
At the top of the report, I saw what concerned my client.
It was this finding in a well-characterized gastric cancer-associated gene called CDH1:
At first glance, this looked very concerning.
Most people have two Gs at this particular location in the CDH1 gene - one G they have received from their mom’s copy of the CDH1 gene, the other G received from their dad’s copy.
My client was reported to have one G and one A at this location, which led it to be red-flagged.
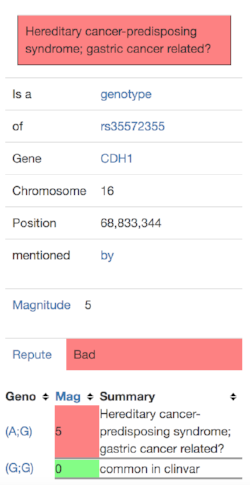
I clicked on the “more info” link and saw this:
This gave me to three questions to investigate further:
Question 1: Is this finding real?
Since both 23andMe and Promethease tell us either in the fine print or during the registration process that we can’t use the information from raw data for medical purposes (because raw data can be wrong - see this Gina Kolata article and Genetics in Medicine peer-reviewed journal article by Tandy-Connor, et al), we would need a clinical test to know for sure whether the discovery of an A instead of a second G represented a real, actual finding or was a result of a technological failure (a misread of the second G as an A). We also would need a clinical laboratory to tell us if this means the particular finding identified is thought to be a disease-causing change (also called pathogenic) or a harmless one (called benign).
Question 2: Let’s assume the finding is real and the client really did have a G change to an A…what does that mean?
The places I turn to next after finding a possibly-important finding from a raw data result are outside databases that allow for further research into individual genetic variants. There are multiple places to go, but the ones I use the most are dbSNP and ClinVar.
Genetic mutations or variations (now more appropriately referred to as variants) are sometimes given a code starting with the letters “rs.” The rs code (or “rsID”) identifies a location for that genetic finding in the genome (the entire stretch of someone’s full DNA sequence, including all of the chromosomes). Different databases are available that track and use that rs code and compile all the information available about that particular variant.
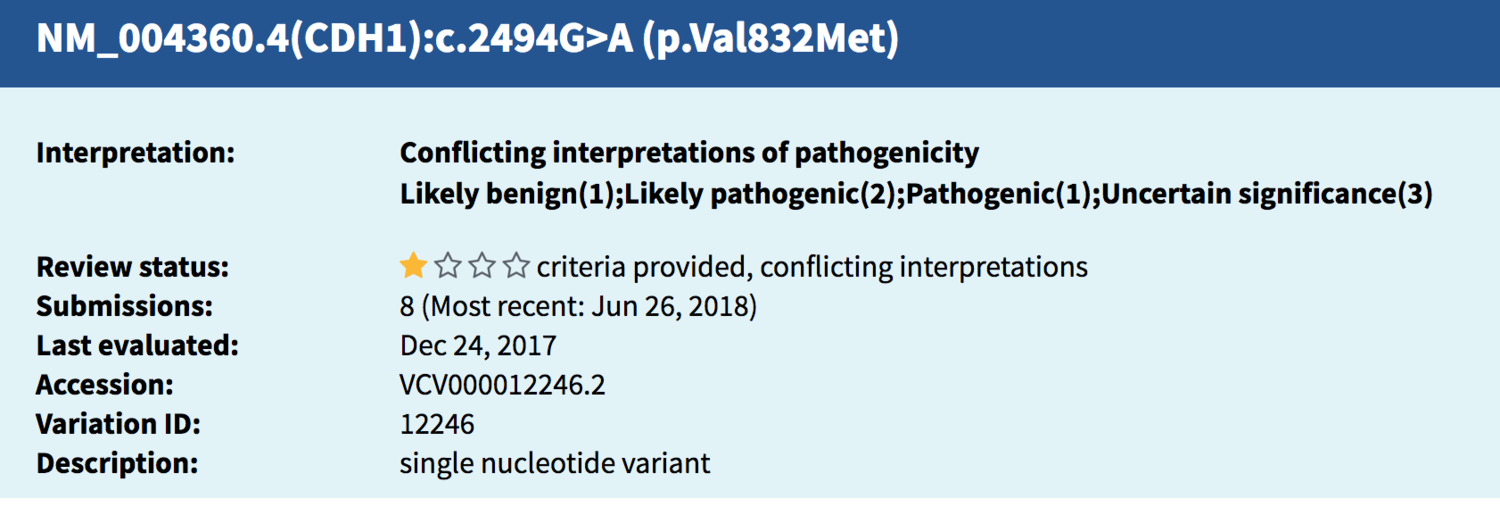
Sometimes, different codes besides an rs number are used to identify the same variant; this particular finding in CDH1 (rs35572355) tracked to this entry (below) found in ClinVar. The string of letters and numbers are identifiers and explain what change has taken place in the DNA from the usual/typical genetic spelling typically seen there.
Source: National Center for Biotechnology Information. ClinVar; Variation ID 12246, https://preview.ncbi.nlm.nih.gov/clinvar/variation/12246 (accessed July 27, 2018).
When I opened up the ClinVar entry for more details, I saw that 8 different submissions had been reported in ClinVar, meaning this same genetic variant reported on my client’s Promethease report had been found in multiple other people in the past. When a laboratory puts information on a variant into ClinVar, it is called a submission, and you can see from the screen shot above the most recent submission occurred in June of 2018.
The ClinVar entry showed that of the 8 submissions, spanning from 2010 to 2018:
One laboratory called this finding likely benign (and I know of a second one who does as well, they aren’t included in the count ClinVar took; more on that later**).
Three labs called it pathogenic or likely pathogenic.
Three labs have the finding labeled as “uncertain,” meaning they haven’t determined yet whether they feel it is benign or pathogenic.
One lab gave no label to the finding at all.
Hm. Well, that’s frustrating. There seemed to be a lack of agreement from one lab to the next whether this was a concerning finding or not.
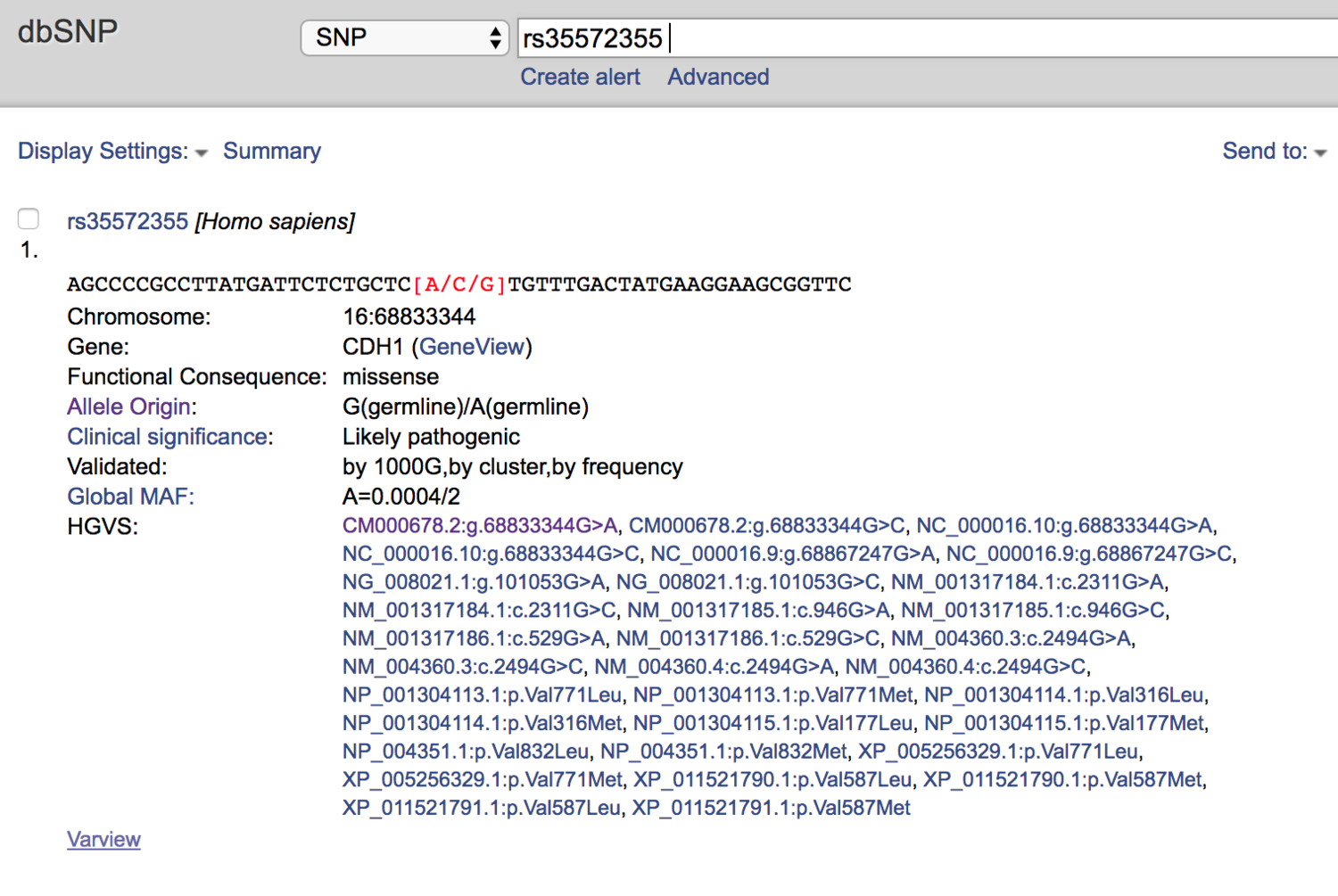
So I turned to dbSNP next.
dbSNP called it “Likely Pathogenic.”
This investigation led me to my third question…
Question 3: Can we get this particular finding retested in a clinical laboratory to find out if it is really there and what it means?
I messaged someone I know at a laboratory that provides clinically-valid testing at the most affordable price I am aware of at this point in time. I gave them the name of the CDH1 variant (that “rs” number) and reported the “genotype call” (remember, this person’s genotype was A;G instead of G;G).
The friendly and helpful genetic counselor I communicated with at the laboratory looked into it for me. She responded quickly, informing me that the testing would not qualify for their low-price “confirmatory testing” option, because they would end up reporting this as a benign finding even if present. (**This step is how I determined there was at least one additional laboratory calling this finding benign but not reported as such in ClinVar.)
So there you have it, folks.
My response to my confused and disappointed client was that even though she had a possibly concerning finding appear in her Promethease report, I still could not recommend she seek out clinical-grade confirmatory testing as a follow up at this point.
Why? Because even if the raw data finding of a G to A change at this particular location in the CDH1 gene were confirmed, it appears that it would be labeled as harmlessly “benign” by some labs and potentially disease-causing “pathogenic” by others.
WHETHER MY CLIENT WOULD BE TOLD SHE CARRIES A DISEASE-CAUSING VARIANT OR A BENIGN ONE WOULD DEPEND ON WHICH LAB SHE SENDS HER DNA SAMPLE TO.
This is not shocking to those of you who work in the medical genetics field. We know it’s rare that this happens, but we know it does. This might come as a shock to those who to this point in time felt medical genetic tests gave a black-and-white, clearly positive or clearly negative result every time.
You might be wondering what I did next. Here were my next-steps:
1. I reached out to Promethease about this case to alert them of the entry and request they update the entry on this particular finding to account for the unusual situation of highly-divergent pathogenicity calls in ClinVar. If this has happened to my client with this particular variant that has now been identified by a number of labs, it will happen again in the future. We need the Promethease report to call this out as a potential false alarm and to report it as a finding for which clinical labs have not reached consensus.
2. I recommended my client NOT do further genetic testing at this time related to this particular finding, and I recommended that instead she:
· research symptoms of gastric cancer of which to be on the lookout.
· consider seeking out and attempting contact with biological family (if she felt ready) in order to pursue family medical history, specifically to see if any gastric cancer history was present; she might also consider contacting the agency who handled her adoption to see if further medical history could be obtained via any connection maintained there.
· consider meeting with a cancer specialist who could determine whether and how to monitor her for gastric cancer symptoms and remain vigilant for other CDH1-related issues until the lack of clarity surrounding this genetic finding is resolved.
Fortunately, this is the most extreme case like it that I have run into. I rarely come across genetic variants that shows such disparate information in ClinVar or other sources of data.
Worded differently, most DNA variants I’ve researched demonstrate consensus from one lab to the next and support the conclusion made by the third-party tool. But you still have to take a deeper look into the supporting data to know for sure.
There were additional complicating factors in this particular case: this client was adopted, and she was of Asian ancestry.
· DNA databases based in the United States (both medical and genealogical) tend to be over-represented with results from those of European ancestry and under-represented by those of other ethnicities. Projects like All of Us are trying to fix that problem, but it is going to take some time before the playing field is leveled.
· When a person is adopted we often have little or no family history information to review. This means we couldn’t check the family history in order to see if anyone was known to have developed gastric cancer or another CDH1-associated cancer in the past, and we couldn’t offer to test other genetic family members. This is one reason of many for my support of adoptee rights to their identity and full history and why you’ll see a lot of adoption-related posts in my Twitter feed and other social media.
In conclusion:
When it comes to raw data, before you accept a finding, try to confirm using multiple sources PLUS a follow-up clinical DNA test.
Yes, it is going to take more time and money. You should know that the next-steps involved in figuring out whether raw data and third party analysis has given you potentially-significant medical information involves spending more time and money and possibly going through a period of anxiety until everything is resolved. If you take the first step with raw data and third-party health tools, you need to prepare yourself for what may come next if you have a worrisome finding appear.
Please help me to spread this message around your online circles and communities. It bothers me when people tell each other to use raw data and an inexpensive third-party tool for medical information without acknowledging any of the short-comings. And it bothers me that when I try to speak up about the short-comings, I’m told I’m being paternalistic.
No DNA test is perfect, but analyses using the raw data from consumer market tests are the weakest of all. I’m not one to stand by and let misinformation be spread without trying to say something about it.
I’m not saying raw data is worthless, I’m just telling you it’s complicated.
Take some time to learn about raw data’s weaknesses and how to overcome them.
Make sure you do your due diligence.